GraSPI functionality¶
GraSPI computes a library of descriptors for a segmented microstructure
and computes descriptors that are relevant for organic solar cells performance.
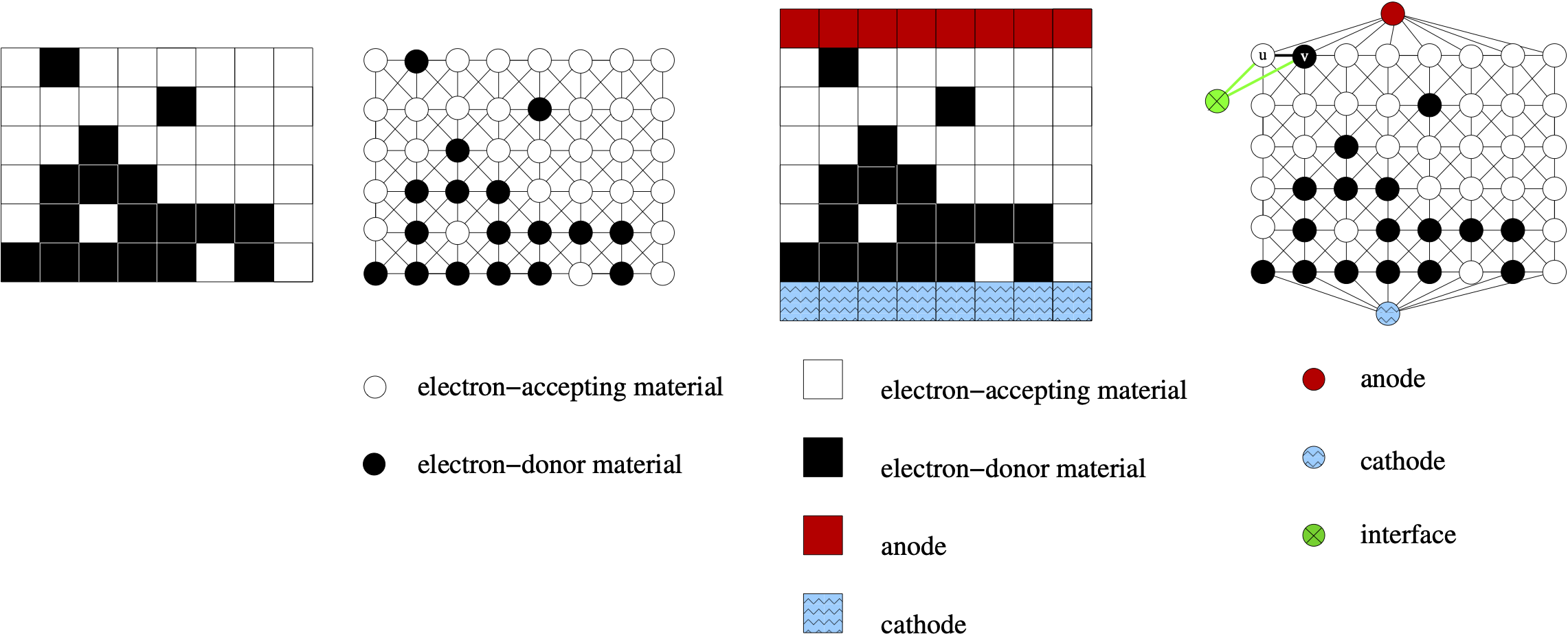
GraSPI assumes that input microstructure is segmented into two phases: BLACK and WHITE.
In our application, organic solar cells, BLACK pixels correspond to electron-donating materials,
and WHITE pixels correspond to electron-accepting material - as marked in the figure below.
Three meta-vertices are added to the network:
BLUE corresponds to cathode,
RED corresponds to anode,
GREEN corresponds to the interface.
As graph is being constructed, the meta-vertices are added to the graph. For the
morphology inputed as an array, the BLUE vertex is added to all vertices in the first row,
and RED vertex is added to all vertices corresponding to the last row in the input file.
GraSPI reads the colors of the pixels starting from the bottom left corner.
GREEN vertices are added as the graph is constructed, when BLACK pixel has any WHITE
voxel in the neighborhood, two additional edges are added (between BLACK and GREEN, and between WHITE and GREEN).
The package computes two types of descriptors: scalar descriptors and array descriptors. The scalar descriptors are directed to the standard output, while array descriptors are directed to the corresponding file. The array descriptors corresponds to the shortest distances and are saved in the following files:
DistancesGreenToRedViaBlack.txt
DistancesGreenToBlueViaWhite.txt
DistancesBlackToRed.txt
DistancesWhiteToBlue.txt
DistancesBlackToGreen.txt
TortuosityBlackToRed.txt
TortuosityWhiteToBlue.txt
The names of the file indicates the conditions used to filter the graph.
For example file DistancesBlackToGreen.txt stores all the shortest distances between any donor/black/0 voxel and the interface/green.
If there is no direct connection between source vertex and the target vertex, the distance is set to inifinity.
This is is how boost library is initializing the distance vector.
The distances are save in the order of the labels from the input file.
The infinity distances are maintained to maintain capability to map back the distances to the input voxels.
GraSPI provides one function where a set of descriptors are computed - see documentation for more details:
-
std::vector<desc_t>
graspi::compute_descriptors_only(int *p_colors, const unsigned int &nx, const unsigned int &ny, const unsigned int &nz = 1, const float &pixelsize = 1.0, const bool &if_per = true) This function computes descriptors of morphology defined on structured matrix
This function computes a set of descriptors of morphology
- Return
the vector of descriptors, where each descriptor is a pair (std::pair<float,std::string>) consisting of descriptor’s value (float) and name (string)
- Parameters
p_colors: is the morphology stored as an array of colors (row-wise order of matrix)nx: is the number of voxels in x-directionny: is the number of voxels in y-directionnz: is the number of voxels in z-direction (default value is 1)pixelsize: is the physical unit of the voxel (distances will be rescaled according to this value)if_per: is the flag setting the periodicity on sides (true value means periodicity on sides, for 2D - in x-dir, for 3D - in x anf y-diretion)